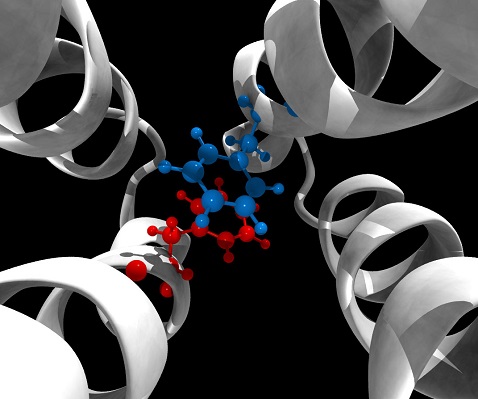
 Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega
Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega Supercomputers Find Bacterial 'Off' Switch
[caption id="attachment_15343" align="aligncenter" width="478"] Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega
Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega
 Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega
Simulation of the molecular 'switch' that tells E. coli to turn itself off.[/caption] The comparatively recent addition of supercomputing to the toolbox of biomedical research may already have paid off in a big way: Researchers have used a bio-specialized supercomputer to identify a molecular "switch" that might be used to turn off bad behavior by pathogens. They're now trying to figure out what to do with that discovery by running even bigger tests on the world's second-most-powerful supercomputer. The "switch" is a pair of amino acids called Phe396 that helps control the ability of the E. coli bacteria to move under its own power. Phe396 sits on a chemoreceptor that extends through the cell wall, so it can pass information about changes in the local environment to proteins on the inside of the cell. Its role was discovered by a team of researchers from the University of Tennessee and the ORNL Joint Institute for Computational Sciences using a specialized supercomputer called Anton, which was built specifically to simulate biomolecular interactions among proteins and other molecules to give researchers a better way to study details of how molecules interact. Anton is a 512-node, built by D.E. Shaw Research and housed at the Pittsburgh Supercomputing Center designed to study the activity, rather than the appearance, of proteins and other bio-molecules known almost exclusively through techniques that produce detailed but static images that show nothing about how the molecules communicate or interact. "For decades proteins have been viewed as static molecules, and almost everything we know about them comes from static images, such as those produced with X-ray crystallography," according to Igor Zhulin, a researcher at ORNL and professor of microbiology at UT, in whose lab the discovery was made. "But signaling is a dynamic process, which is difficult to fully understand using only snapshots." Rather than sitting still on the tip of an E. coli chemoreceptor, Phe396 flips 180 degrees back and forth, apparently turning itself and the motility mechanism it helps direct on and off. A paper describing the discovery was published Dec. 13 in the journal Nature Communications. Zhulin's team couldn't observe Phe396's acrobatics directly, but were able to identify its behavior by running a simulation on Anton of the interaction of the 140,000 atoms that make up the motor-control-signaling portion of the chemoreceptor. Because it was designed and programmed specifically for that type of simulation, Anton is able to run molecular-dynamic simulations almost two orders of magnitude faster than any other system available in 2010, when it went online, according to the National Resource for Biomedical Supercomputing. In 2010, it simulates changes in the three-dimensional structure of a protein molecule in a millisecond – a hundred times faster than the previous record, according to a 2010 article in the journal Nature. The result "exemplifies the growing importance of numerical experiments in biology," according to statements from another team member, Jerome Baudry, assistant professor in the UT biochemistry and molecular biology departments. The 140,000 atoms at the tip of the chemoreceptor are only the tip of the iceberg, however. Phe396 consists of a pair of identical molecules, but is grouped with two other sets of pairs, each of which are grouped in threes to form larger signaling units, which could amplify, modify or nullify signals from the first pair. Confirming how all those molecules work together requires a simulation of almost 400,000 atoms and the increasingly complex physics of their interactions. "Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan." Titan is a Cray, Inc. XK7 supercomputer installed at Oak Ridge National Laboratories with a peak performance of 20 petaflops, or 20,000 trillion calculations per second. Until it was edged out in June – by Tianhe-2, a Chinese behemoth with 16,000 nodes, 3,120,000 computing cores and a top sustained speed of 33.86-petaflops – Titan was the fastest supercomputer in the world. "With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines." With a good-enough idea of the language of those signals and the cell's communication pathways, it might be possible to create treatments that simply tell pathogens to turn themselves off or stop activity causing damage to the rest of the body, without resorting to poisons, birth control, artificially super-powering the antibody system or any of the other techniques custom-designed medicines use to block or slow down activity in the absence of a clear stop sign written in the pathogen's own language. Image: UT/David Ortega 

